Walkthrough of Nested Sampling#
We walk through the same components in the Quickstart, to give more detail as to what the choices mean.
Problem definition#
We show how to use the basic user interface for Nested Slice Sampling. We will set up a 20D problem involving a multivariate unit Gaussian \(\mathcal{N}(0,\mathbf{I})\) prior, and a multivariate Gaussian likelihood with a randomized mean and covariance. The resulting posterior and normalizing constant is analytically known.
import jax
import jax.numpy as jnp
import tqdm
import blackjax
rng_key = jax.random.PRNGKey(0)
d = 20
C = jax.random.normal(rng_key, (d, d)) * 0.1
like_cov = C @ C.T
like_mean = jax.random.normal(rng_key, (d,))
prior_mean = jnp.zeros(d)
prior_cov = jnp.eye(d) * 1
Components needed#
In a similar style to Sequential Monte Carlo methods available within blackjax, We need to declare three pieces to compose the sampling algorithm
A (log) prior density
A (log) likelihood function
An initial population of particles, drawn from the prior. This will remain fixed throughout a run, so the size of this population is a hyperparameter of the algorithm, we will call this
n_live
logprior_density = lambda x: jax.scipy.stats.multivariate_normal.logpdf(
x, prior_mean, prior_cov
)
loglikelihood_function = lambda x: jax.scipy.stats.multivariate_normal.logpdf(
x, like_mean, like_cov
)
rng_key, prior_sample_key = jax.random.split(rng_key)
n_live = 1000
initial_population = jax.random.multivariate_normal(
prior_sample_key, prior_mean, prior_cov, (n_live,)
)
Algorithm definition#
We have already implicitly set one hyperparameter we need, n_live, the remaining ones to set are
num_inner_steps– the length of the short Markov chains used to update the population, typically set to some multiple of the dimension of the parameter space, \(3 \times d\) is a good starting pointnum_delete– some integer \(\in [1, n_\mathrm{live} -1]\), how many chains to vectorize over, setting this to approximately 10% ofn_liveis likely good for CPU usage, Up to 50% for GPU.
nss is defined as a top level API in the blackjax library, initialising this function returns a blackjax.SamplingAlgorithm. This base type defines the two pieces needed to build a sampling algorithm,
init– A function to initialiseNSStatefrom particle positionsstep– A function to evolve theNSStatepopulation one step
num_inner_steps = 3 * d
num_delete = int(n_live*0.1)
algo = blackjax.nss(
logprior_fn=logprior_density,
loglikelihood_fn=loglikelihood_function,
num_delete=num_delete,
num_inner_steps=num_inner_steps,
)
state = algo.init(initial_population)
The state is constructed as a NamedTuple, consisting of the following fields
from blackjax.ns.base import NSState
print(f"{NSState.__doc__}")
State of the Nested Sampler.
Attributes
----------
particles
A PyTree of arrays, where each leaf array has a leading dimension
equal to the number of live particles. Stores the current positions of
the live particles.
loglikelihood
An array of log-likelihood values, one for each live particle.
loglikelihood_birth
An array storing the log-likelihood threshold that each current live
particle was required to exceed when it was "born" (i.e., sampled).
This is used for reconstructing the nested sampling path.
logprior
An array of log-prior values, one for each live particle.
pid
Particle ID. An array of integers tracking the identity or lineage of
particles, primarily for diagnostic purposes.
logX
The log of the current prior volume estimate.
logZ
The accumulated log evidence estimate from the "dead" points .
logZ_live
The current estimate of the log evidence contribution from the live points.
inner_kernel_params
A dictionary of parameters for the inner kernel.
Iteratively updating the state using algo.step is a nested sampling algorithm. To do parameter estimation we need to track what we left behind, another NamedTuple this time NSInfo. The quantity we read out is the accumulation of many NSInfo tuples associated with each state transition. The NSInfo consists of the following fields:
from blackjax.ns.base import NSInfo
print(f"{NSInfo.__doc__}")
Additional information returned at each step of the Nested Sampling algorithm.
Attributes
----------
particles
The PyTree of particles that were marked as "dead" (replaced) in the
current step.
loglikelihood
The log-likelihood values of the dead particles.
loglikelihood_birth
The birth log-likelihood thresholds of the dead particles.
logprior
The log-prior values of the dead particles.
inner_kernel_info
A NamedTuple (or any PyTree) containing information from the update step
(inner kernel) used to generate new live particles. The content
depends on the specific inner kernel used.
Following the general design pattern of blackjax, we set up a compilable one_step wrapping around the algo.step.
@jax.jit
def one_step(carry, xs):
state, k = carry
k, subk = jax.random.split(k, 2)
state, dead_point = algo.step(subk, state)
return (state, k), dead_point
If we know how many steps to take we can now jax.lax.scan over this, compose some larger number of steps etc. As we won’t in general know how many steps we need, and we need to save every NSInfo, we will use a python outer loop. Any inefficiency of this can be lifted by scanning over some multiple of one_step to build a coarser step in python.
A nested sampling run#
We will assess whether we have run enough steps of sampling dynamically by checking some properties of the sample state. In this example we will follow conventions of the PolyChord implementation [HHL15], and check when the remaining live evidence is less than \(1e^{-3}\) of the running estimate.
dead = []
with tqdm.tqdm(desc="Dead points", unit=" dead points") as pbar:
while not state.logZ_live - state.logZ < -3:
(state, rng_key), dead_info = one_step((state, rng_key), None)
dead.append(dead_info)
pbar.update(num_delete)
Dead points: 41500 dead points [00:11, 3713.17 dead points/s]
Utilities#
As the calculation is a little unusual, paying little attention to the final state, and more to the discarded infos, we include some utils to do common manipulations
finalise– For correctness the final state should be “killed” and rendered as an info, a crude function to do this compresses the list ofdeadand the finalstateinto a single cohesiveNSInfolog_weights– compute posterior point weights fromNSInfo, this should reconstruct the normalising constant, typically used in conjunction withfinalisesample– draw “equal weight” posterior samples fromNSInfoess– simply get an estimate of the Effective Sample Size, useful to know how many posterior points you can roughly usefully sample
from blackjax.ns.utils import log_weights, finalise, sample, ess
rng_key, weight_key, sample_key = jax.random.split(rng_key,3)
final_state = finalise(state,dead)
log_w = log_weights(weight_key, final_state, shape=100)
samples = sample(sample_key, final_state, shape = n_live)
ns_ess = ess(sample_key, final_state)
logzs = jax.scipy.special.logsumexp(log_w, axis=0)
We can compare our estimate of the normalizing constant and use the samples to construct any useful posterior estimates
from jax.scipy.linalg import solve_triangular, cho_solve
def compute_logZ(mu_L, Sigma_L, logLmax=0, mu_pi=None, Sigma_pi=None):
"""
Compute log evidence, posterior covariance, and posterior mean.
Args:
mu_L: Likelihood mean
Sigma_L: Likelihood covariance
logLmax: Maximum log likelihood value
mu_pi: Prior mean
Sigma_pi: Prior covariance
Returns:
Tuple of (log evidence, posterior covariance, posterior mean)
"""
# Use Cholesky decomposition for more stable calculations
L_pi = jnp.linalg.cholesky(Sigma_pi)
L_L = jnp.linalg.cholesky(Sigma_L)
# Compute precision matrices (inverse covariances)
prec_pi = cho_solve((L_pi, True), jnp.eye(L_pi.shape[0]))
prec_L = cho_solve((L_L, True), jnp.eye(L_L.shape[0]))
# Compute posterior precision and its Cholesky factor
prec_P = prec_pi + prec_L
L_P = jnp.linalg.cholesky(prec_P)
# Compute Sigma_P using Cholesky factor
Sigma_P = cho_solve((L_P, True), jnp.eye(L_P.shape[0]))
# Compute mu_P more stably
b = cho_solve((L_pi, True), mu_pi) + cho_solve((L_L, True), mu_L)
mu_P = cho_solve((L_P, True), b)
# Compute log determinants using Cholesky factors
logdet_Sigma_P = -2 * jnp.sum(jnp.log(jnp.diag(L_P)))
logdet_Sigma_pi = 2 * jnp.sum(jnp.log(jnp.diag(L_pi)))
# Compute quadratic forms using Cholesky factors
diff_pi = mu_P - mu_pi
diff_L = mu_P - mu_L
quad_pi = jnp.sum(jnp.square(solve_triangular(L_pi, diff_pi, lower=True)))
quad_L = jnp.sum(jnp.square(solve_triangular(L_L, diff_L, lower=True)))
return (
(
logLmax
+ logdet_Sigma_P / 2
- logdet_Sigma_pi / 2
- quad_pi / 2
- quad_L / 2
),
Sigma_P,
mu_P,
)
log_analytic_evidence, post_cov, post_mean = compute_logZ(
like_mean,
like_cov,
mu_pi=prior_mean,
Sigma_pi=prior_cov,
logLmax=loglikelihood_function(like_mean),
)
print(f"ESS: {int(ns_ess)}")
print(f"logZ estimate: {logzs.mean():.2f} +- {logzs.std():.2f}")
print(f"analytic logZ: {log_analytic_evidence:.2f}")
ESS: 9875
logZ estimate: -32.75 +- 0.19
analytic logZ: -32.83
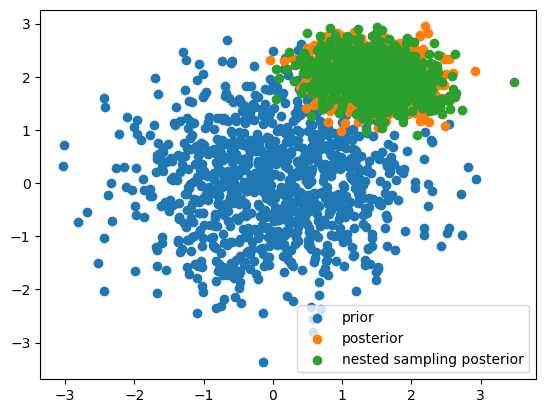
Lastly we can use the particles themselves to do parameter inference, lets look at the first corner of our 20D parameter space
import matplotlib.pyplot as plt
posterior_truth = jax.random.multivariate_normal(
rng_key, post_mean, post_cov, (n_live,)
)
plt.scatter(
initial_population[..., 0], initial_population[..., 1], label="prior"
)
plt.scatter(
posterior_truth[..., 0], posterior_truth[..., 1], label="posterior"
)
plt.scatter(
samples[..., 0], samples[..., 1], label="nested sampling posterior"
)
plt.legend()
<matplotlib.legend.Legend at 0x379add400>